Self-Supervised Learning
Machine learning promises automation. In an era where we can generate millions of images in a day, we’d really like a computer to be able to look at these images for us and identify which ones are most interesting. But in practice, to implement machine learning, users either have to label potentially millions of images to train a neural network, so it’s not much less effort than just looking at screens manually.
To address this issue, we use self-supervised learning. Self-supervised learning methods are inspired by developmental psychology. Unlike state-of-the-art machine learning methods, which rely on more rote learning strategies, children learn skills through play and exploration. Similarly, training neural networks on autonomous puzzle-solving tasks can help them develop skills and representations that can be transferred to more useful analyses. By carefully designing the task to recognize specific biology in images, we can control what the model learns.
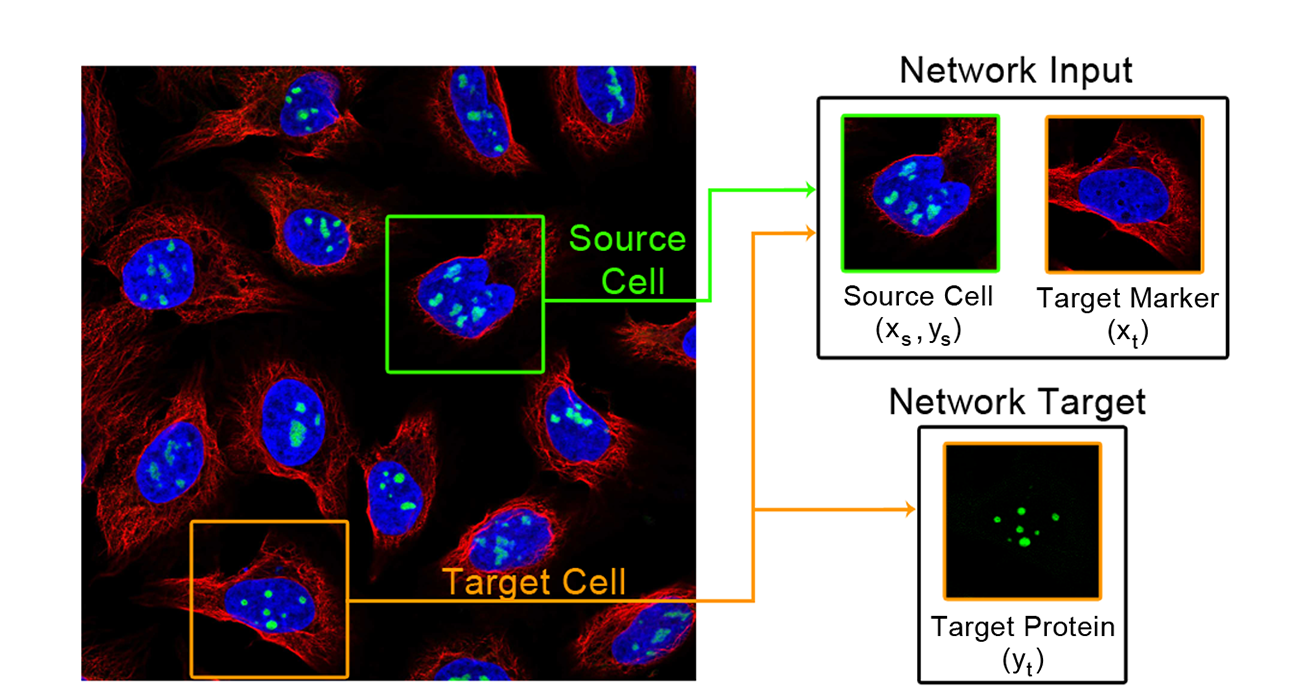
For microscopy images, we designed a task called “paired cell inpainting". We ask the model to solve a “coloring book” challenge: the task asks it to recognize protein localization in one cell, so it can color in the right organelle in a second different cell. Since this task implicitly requires an understanding of protein localization in the first cell, this representation can be transferred to downstream analyses. Our representations perform better than those laboriously designed by experts in classifying protein subcellular localization, suggesting that they capture more biology. In addition, our method is general: we were able to analyze datasets that were never analyzed computationally previously due to technical challenges.
We also applied these methods to the intrinsically disordered “dark proteome”. These are regions of proteins that don’t fold into a stable secondary/tertiary structure. Although they are widespread and critical to protein function, we still don’t understand them well because they evolve too rapidly to be analyzed by classic bioinformatics methods. By creating a self-supervised method, “reverse homology", that exploits principles of evolutions to learn about conserved elements of these sequences, we can automatically learn hundreds of important features that must be conserved over evolution for these regions of proteins to carry out their function. Interpreting these features lets us produce hypotheses that fuel biological discovery.